Brain cancers
PhD project
Introduction
Cells grow in an environment that influences them. This is especially true for tumor cells, which, as part of an ecosystem, are influenced by interactions with other tumor cell types and with the tumor microenvironment. Detailed molecular characterization of cell types composing glioma brain tumors has been performed but a lot less is known regarding the influence of the cell types on each other and how this contributes to tumor progression. With this PhD project, we aim to decipher how the tumor cell types influence each other, how they interact with neurons, and how these interactions contribute to tumor progression through specific transcriptional programs. We will especially focus on the interactions between different glioma tumor cell types, and tumor cells and neurons, for which important key aspects still need to be discovered.
Spatial heterogeneity of IDH-mutant gliomas
This project focuses on three types of glioma: glioblastoma (GBM) the most common and aggressive adult brain tumor with a median survival of approximately 15 months and an extremely poor prognosis despite aggressive therapies, and two types of high-grade isocitrate dehydrogenase (IDH)-mutant gliomas, oligodendrogliomas, and astrocytoma, occurring in young adults which also represent an important clinical problem due to high rate of recurrence leading to a fatal issue. We will first analyze novel spatial transcriptomics data recently generated by our collaborator with 10X Visium technology from human glioma samples with a special focus on (i) the core of the tumor with dense tumor cells content, and (ii) areas of perineuronal satellitosis where clusters of glioma cells surround neurons cell bodies, to reveal the gliomas’ subtype spatial organization.
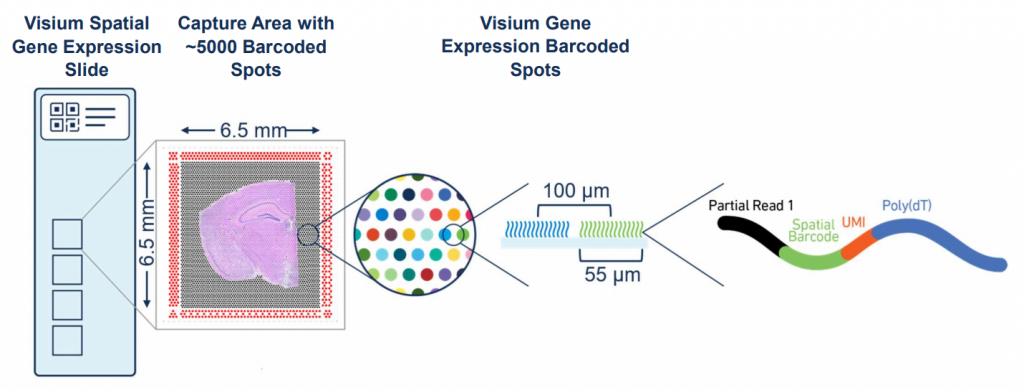
Using up-to-date methods for spatial transcriptomics such as Loupe (10X Genomics viewer) and Cell2Location (spatial deconvolution) within R/Python environment, we will identify the tumor cell types’ spatial localization. This will give us access to unique spatial maps of these gliomas and allow us to compare them as well as identify the cells states that are in contact to each other’s. Then, we will decipher the tumor cell-cell interactions based on our identified tumor cell types in contact, Ligand-Receptors databases and network methods (eg., CellTalkDb, NicheNet). We will be able to link cells interactions to transcriptional programs and thus infer the effects of such interactions on regulatory pathways and their contribution to tumor progression in each glioma subtype.
Crosstalks between neurons and tumor cells
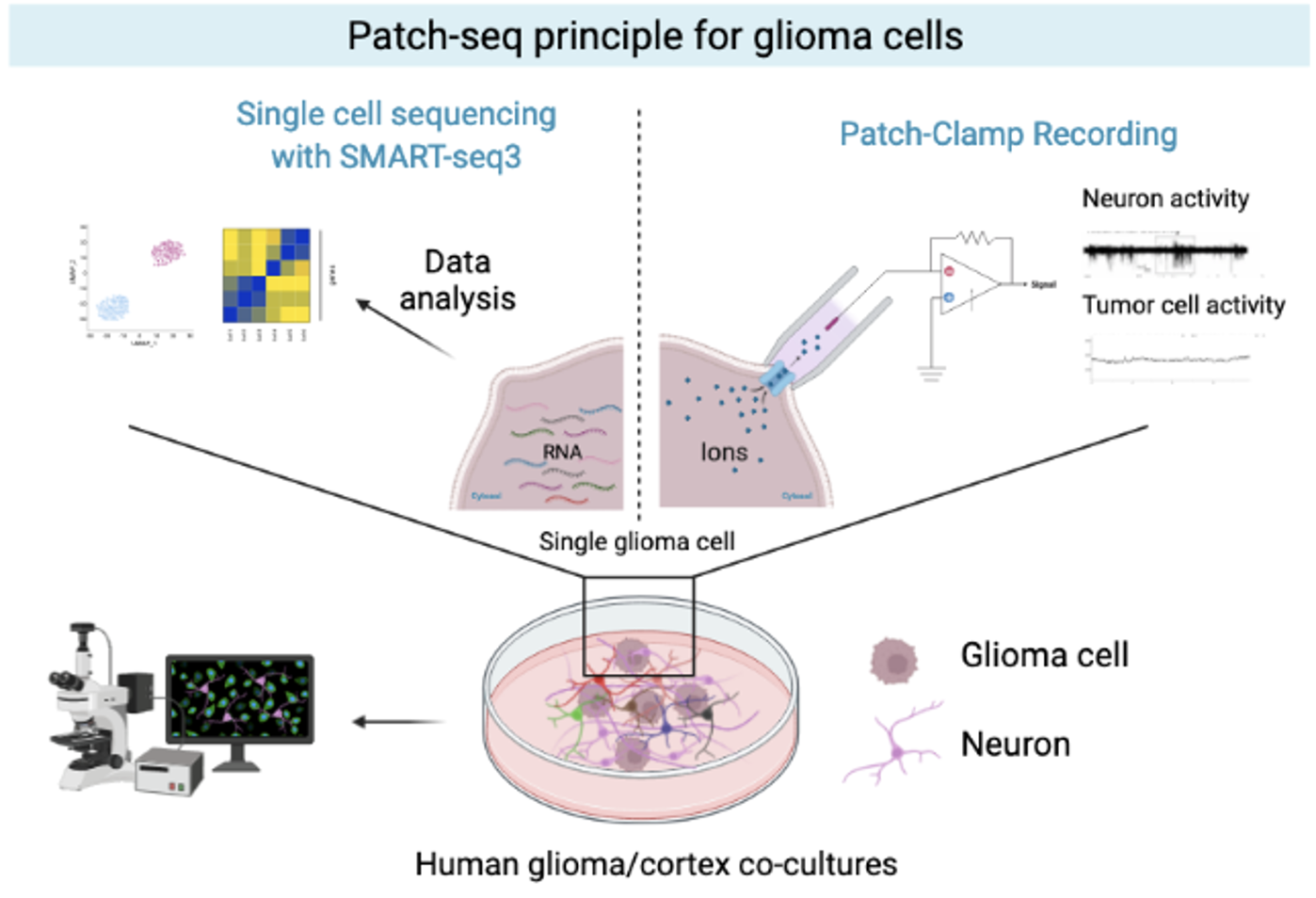
In the second part of the PhD, we aim to further characterize the tumor cell-neuron interaction. From cortex-glioma co-cultures performed by our collaborator, we will study the effect of normal tissue interaction on glioma transcriptome by bulk RNA-seq. Then, together with our collaborator, we will generate a new Patch-seq data type from tumor cells following neuronal activation or not. Patch-seq experiment is a combination of patch experiment and single-cell RNA-seq with SMART-seq3 allowing a morphological, electrophysiological, and single-cell transcriptional profiling characterization of a given cell. We will therefore characterize the features of tumor cells receiving a neuronal synapse and decipher tumor cell-neuron interactions following neuronal activation and their effect on the transcriptional program driving tumor growth. Overall, this PhD project will provide novel biological insights into functional glioma tumor heterogeneity, a new mechanistic understanding of tumor cell interactions, and their effect on tumor progression. It will also highlight the heterogeneity between the glioma subtypes and provide useful appreciations of their differences. Ultimately, our findings could lead to further development of pre-clinical models and novel rationales for therapeutic targeting.
Milestone of the project
- September 2022 - Best Poster Award at 5th Course on Computational Systems of Biology of Cancer, Institute Curie, Paris, France
- September 2024 - Best Poster Award at the Oligodendrogliomas Workshop organized by the POLA network
- February 2025 - Our review untitled Cell-type deconvolution methods for spatial transcripotmics has been accepted for publication in Nature Reviews Genetics. We provide a comprehensive landscape of cell-type deconvolution and created a web-based tool to ease method selection and development.
Poster and talks related to the project
- September 2022 - Best Poster Award at 5th Course on Computational Systems of Biology of Cancer, Institute Curie, Paris, France
- September 2023 - Poster presentation at 6th Course on Computational Systems of Biology of Cancer, Institute Curie, Paris, France
- May 2024 - Poster presentation at International Curie Symposium, Institute Curie, Paris France
- June 2024 - Poster presentation at VIB – Spatial Omics Conference, Ghent, Belgium
- September 2024 - Poster presentation at the Oligodendrogliomas Workshop organized by the POLA network
- January 2025 - Poster presentation at Brain Tumor microenvironment symposium, Paris, France


